Projects
Functional Analysis of Influenza Virus PA-X and NS1 in Host Shutoff
 Influenza virus infection causes global inhibition of host protein synthesis in infected cells. This host shutoff is thought to allow viruses to escape from the host antiviral response, which restricts virus replication and spread. The virus expresses two accessory proteins NS1 and PA-X to induce general shutoff. We analyze the molecular mechanism of PA-X-induced host shutoff, functional interplay between PA-X and NS1, and their role in regulation of host innate and acquired immune responses.
Influenza virus infection causes global inhibition of host protein synthesis in infected cells. This host shutoff is thought to allow viruses to escape from the host antiviral response, which restricts virus replication and spread. The virus expresses two accessory proteins NS1 and PA-X to induce general shutoff. We analyze the molecular mechanism of PA-X-induced host shutoff, functional interplay between PA-X and NS1, and their role in regulation of host innate and acquired immune responses.
Learn more about Functional Analysis of Influenza Virus PA-X and NS1 in Host Shutoff
Molecular Mechanism of Paramyxovirus Assembly
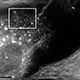 Human parainfluenza virus type 1 (hPIV1) and its counterpart, Sendai virus (SeV; murine parainfluenza virus type 1) are respiratory pathogens in the Paramyxoviridae family, which includes many clinically important human pathogens. We study the mechanism of how viral structural components are assembled in infected cells. This project aims to unveil the molecular interactions between viral and host proteins allowing efficient assembly and release of progeny virions, and the role of cholesterol in virus stability, transmission and infection.
Human parainfluenza virus type 1 (hPIV1) and its counterpart, Sendai virus (SeV; murine parainfluenza virus type 1) are respiratory pathogens in the Paramyxoviridae family, which includes many clinically important human pathogens. We study the mechanism of how viral structural components are assembled in infected cells. This project aims to unveil the molecular interactions between viral and host proteins allowing efficient assembly and release of progeny virions, and the role of cholesterol in virus stability, transmission and infection.
Learn more about Molecular Mechanism of Paramyxovirus Assembly
Role of Influenza Polymerase in Host Adaptation
 Influenza A viruses infect a wide range of hosts, including humans, swine and avian species. Most avian viruses do not infect or replicate well in human hosts. However, some mutations in viral proteins drastically change the host range of the virus. We analyze the mechanism of how mutations in viral polymerase change the host range of the virus.
Influenza A viruses infect a wide range of hosts, including humans, swine and avian species. Most avian viruses do not infect or replicate well in human hosts. However, some mutations in viral proteins drastically change the host range of the virus. We analyze the mechanism of how mutations in viral polymerase change the host range of the virus.
Learn more about Role of Influenza Polymerase in Host Adaptation