URMC / Labs / O'Connell Lab / Projects / The development of RNA-targeting CRISPR tools to study RNA-mediated gene regulation
The development of RNA-targeting CRISPR tools to study RNA-mediated gene regulation
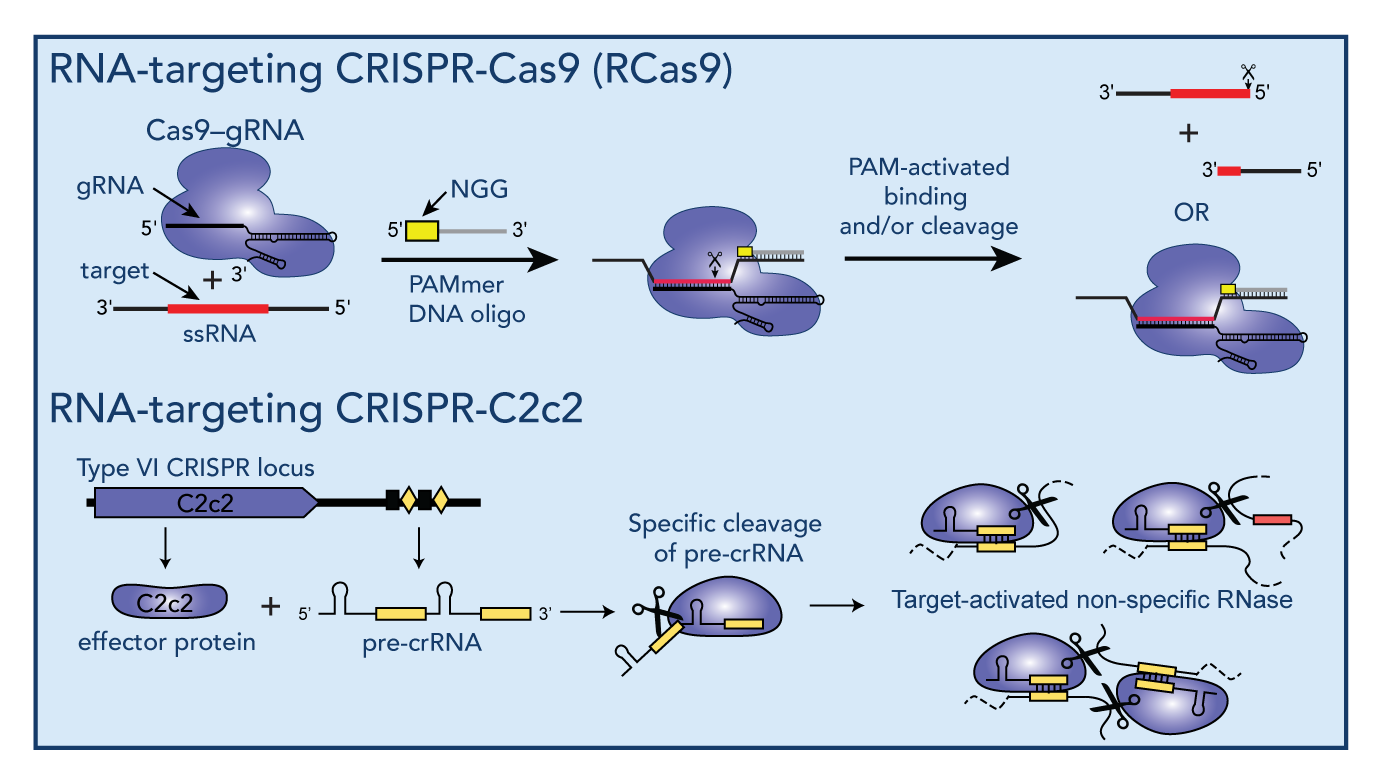
Readily-programmable RNA-targeting CRISPR/Cas systems have recently revealed new opportunities to create a versatile range of tools designed to modulate RNA splicing, RNA translation, RNA editing or to visualize RNAs in live cells. During my postdoctoral research, I developed a versatile tool based on the genome-editing reagent CRISPR/Cas9 to specifically bind and/or cleave any desired RNA sequence in a readily programmable manner (RCas9), which has led to the development of a number of new Cas9 applications including tagless live-cell RNA-imaging and tagless RNA isolation. More recently, we have begun to explore the RNA-targeting abilities of members of the newly-discovered class VI CRISPR system family, and demonstrated that C2c2, an RNA-targeting member of this family has dual-RNase activities that allow both guide-RNA processing as well as target-activated general RNA degradation.
The O’Connell lab has a number of ongoing projects in the lab and with collaborators to continue developing tools CRISPR tools for a range of RNA-targeting applications. Our ultimate aim is to use these tools to not only help unlock new areas of enquiry into interesting facets of RNA biology but also to develop new therapeutic avenues for treating diseases where normal RNA metabolism has been dysregulated.