EHR Data for Research
EHR Data for Research
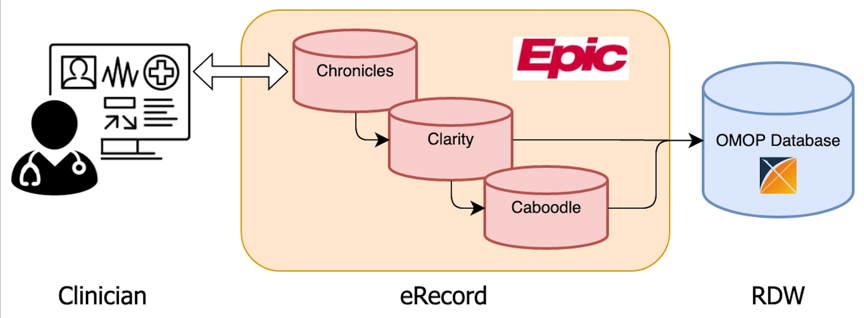
University of Rochester researchers can request clinical data for IRB-approved research projects or Preparatory to Research projects from UR CTSI Informatics and Analytics. Requests can be fulfilled directly from EPIC, or, for faster service, from the URMC OMOP Database. The OMOP database collects clinical data from eRecord in support of clinical and translational research. The database is built upon the Observational Medical Outcomes Partnership (OMOP) Common Data Model to provide a research friendly and open community data standard, and to facilitate optimal data sharing and interoperability with internal and external research partners.
Features
The URMC OMOP Database establishes a common data standard and enables our team to provide the following types of data to researchers much faster:
- Protected Health Information (date of birth, zip code, dates of clinical events)
- Demographics (race, gender, ethnicity, primary care provider)
- Death (date of death, primary cause)
- Vital signs (height, weight, BMI, temperature, blood pressure, pulse, FiO2, respirations, and head circumference)
- Diagnoses (encounter diagnoses, problem lists, medical histories, professional charge transaction diagnoses, final coded diagnoses)
- Chief complaints (reason for visit)
- Procedures (Surgeries, clinical procedures, and procedures on billing)
- Visits (outpatient visits, ED visits, inpatient admissions including ICU stays, telemedicine)
- Medication orders (inpatient medication administration, outpatient drug prescriptions)
- Laboratory tests results
- Social determinants of health (food, housing, transportation, stress, financial strain, partner violence, social connection)
- Social history (smoking, alcohol consumption)
- Primary insurance coverage
- Locations/departments where healthcare is delivered
- Providers (specialties, departments)
- Allergy
- MyChart Activation
- Patient report outcomes (PROMIS survey)
- Research contact preference
- Immunization
- Cancer Staging
- Surgical Procedure
- RUCA code
Data in the OMOP database is loaded from over 2 million patients from eRecord for the following UR Medicine facilities:
- Strong Memorial Hospital
- Highland Hospital
- FF Thompson Hospital
- UR Medicine Ambulatory Clinics
- URMC Urgent Care Centers
- Jones Memorial Hospital (as of 3/5/2022)
- Noyes Memorial Hospital (as of 3/5/2022)
- St. James Hospital (as of 3/5/2022)
Get Started
To get started, please request data through a CTSI Informatics Service Request.
Resources
FAQs
Why OMOP?
The OMOP common data model has been widely adopted by the research community, including the National COVID Cohort Collaborative, due to its versatility and robust features. The benefits of having the OMOP data model implemented at URMC include:
- Patient-centric model with standardized vocabularies and standard data structure
- Standardized data model supports faster return of data extracts for research projects
- Enables open-source tools for data quality assessment, integration of natural language processing (NLP) for patient phenotyping, etc.
- Increased data interoperability facilitates collaborative network studies
- OMOP is increasingly preferred as the “data standard” for external research collaborations.
How It Works
Clinical data is extracted from the Epic Caboodle and Clarity databases of eRecord, transformed into the OMOP Common Data Model format, and loaded into to the OMOP database. The data is updated on a monthly basis.