Structural Biology Services
Crystal Screening
As a preface to structure determination, we offer screening of proteins, DNA, RNA or complexes thereof in a high-throughput sparse matrix format. The facility is equipped with a Mosquito™ liquid handling robot (TTP labtech) capable of establishing 96 screening trials in a matter of minutes. A minimum volume of 25 L of pure concentrated sample (5-10 mg mL-1) is required to establish 96 screens, each in a total initial volume of 400 nL. Dr. Jenkins will train all first-time users, but subsequent trials can be conducted on an independent basis. Experiments will be housed by the facility in a temperature-controlled incubator (20 °C or 4 °C available). Clients are responsible for documenting the results of their own crystallization trials by light microscopy, but will receive initial instructions by Dr. Jenkins. Trials will be discarded after 2 months. Suitable crystals can be harvested and processed for X-ray diffraction experiments.
X-ray Diffraction Crystal Screening
Once a suitable crystal has been grown, it can be mounted for X-ray diffraction analysis to establish whether it is a macromolecule (not salt), its diffraction resolution, and possible unit cell dimensions. First-time users can be trained in this methodology including the use of Proteum© data analysis software (Bruker AXS). As a matter of preference, the entire process of crystal screening can be performed on a fee-for-service basis. The actual process of screening will take 1-2 hr depending on the need to preserve the crystal by flash cooling and subsequent storage in liquid N2 at -178 °C.
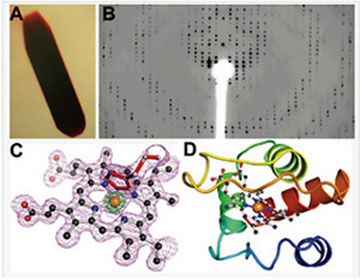
The major steps of an X-ray-crystallographic structure determination. (A) Crystals of cytochrome c552 grown from the Mosquito robot. (B) Representative X-ray diffraction pattern from a CCD detector. (C) Electron density (mesh) for the heme center of a representative cytochrome at 1.4 Å resolution. (D) A backbone ribbon diagram of the protein structure with the heme shown as a ball-and-stick model derived from the high resolution electron density. (Data courtesy of the Dept. of Chemistry, University of Rochester).
X-ray Diffraction Data Collection and Data Processing
We offer the ability to collect a complete X-ray diffraction data set on crystals. This work can be a continuation of X-ray diffraction screening or planned use based on prior characterization. The goal of this work is to obtain a complete, 3-4-fold redundant data set suitable for space group characterization and possible molecular structure determination. Dr. Jenkins can train first-time users in this technique and in use of data reduction software. This aspect of the facility can be performed on a fee for service basis as well. This process may take 4-20 hr depending on the crystal characteristics (size, symmetry, signal-to-noise). Crystals requiring excessive data collection times will be recommended for synchrotron radiation data collection time. Additional time may be necessary to preserve the crystal by transfer from the goniometer head to a liquid nitrogen storage dewar at -178 °C.
Synchrotron-Radiation X-ray Data Collection
Crystals that are poorly matched for the intensity requirements of the in-house X-ray system may be candidates for X-ray data collection at a synchrotron source. We offer routine data collection at such resources, but complex phasing experiments (such as MAD phasing) will require independent time requests or user arrangements with synchrotron sources due to the demanding nature of these experiments. The turn around time for such data is 3-4 months. Users will be instructed in data processing techniques, or can request data processing by Dr. Jenkins on a fee-for-service basis.
X-ray Crystal Structure Determination
We offer assistance with structure determinations by difference Fourier and molecular replacement methods. We offer training in the refinement of crystallographic models against X-ray data as well. Alternatively, Dr. Jenkins can determine and refine structures on a fee-for-service basis. Such experimental structure determinations are arranged on an individual basis. Due to the potentially time-consuming nature of such projects, it will be necessary to consult with the Executive Director about cost-sharing on a 'fee-for-service' versus a 'key personnel' basis (e.g. underwritten by grant support).
Cryo-Electron Microscopy (cryo-EM) Structure Determination
Cryo-EM has quickly become the go-to technology for determining the 3D structures of large macromolecules. URMC has acquired a Thermo Fisher Talos L120C electron microscope to be used as at screening tool to evaluate sample preparations and generate low resolution structures before sending a sample off to a facility with a more powerful system. Dr Jenkins is available to help researchers with design and application of purification procedures to prepare macromolecules for use in cryo-EM. We offer training and assistance using the Vitrobot Mark IV for sample grid preparation. Once vitrified, sample grids can be safely stored in liquid nitrogen dewars located in SBBF or transported via SBBF’s dry-shipper/cryo-dewars to the in-house Electron Microscopy Core or other cryo-EM facilities for grid screening and data collection. We also offer researchers help with cryoEM image processing, 3D model reconstruction, refinement and validation using computing resources available through CIRC and the SBFF.
Homology Modeling (And Notes About Docking)
We offer advice on modeling protein structures, but are primarily an experimental facility. In cases where an experimental structure is known at atomic resolution (i.e. present in the Protein Data Bank), Dr. Jenkins offers advice on rudimentary structure-based experiments, such as point mutations. For cases where a structure of a homologue with >30% sequence identity is known, we can advise individuals on the use of external resources such as SWISS-MODEL, MOD-WEB, or stand-alone packages such as MODELLER. Due to the non-experimental nature of computational modeling, we cannot guarantee the accuracy of modeling results. At present, we do not support molecular docking or virtual screening of diversity libraries. Several good programs are available such as AUTODOCK and the Docking Server. However, we can provide limited advice about these programs to get users started on their own independent projects.
Offline Software Installation
We also offer off-line software installation assistance for any persons wishing to use associated licensed software on their own computers. PC and Mac operating systems only.