Use of Reverse Genetics to Study Influenza Virus Tropism in vivo with a Pandemic Strain of Influenza
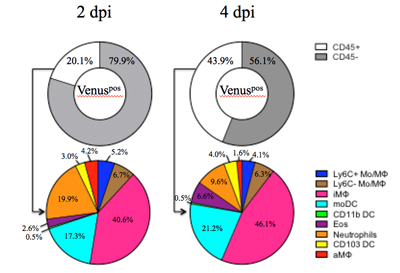
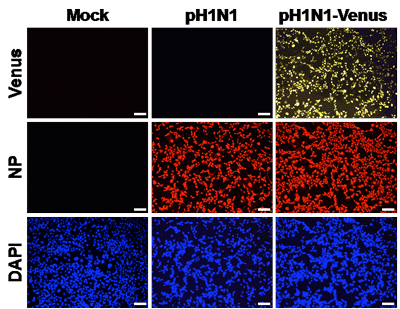 Although it is well established that influenza A virus infection is initiated in the respiratory tract, the sequence of events and the cell types that become infected or access viral antigens remains incompletely understood. In collaboration with Dr. Luis Martinez, we have created and studied a novel Influenza A/California/04/09 (H1N1) reporter virus that stably expresses the Venus fluorescent protein. This has allowed us to identify antigen-bearing cells over time in a mouse model of infection using flow cytometry. Thus far, our studies have revealed that many hematopoietic cells, including subsets of monocytes, macrophages, dendritic cells, neutrophils and eosinophils acquire influenza antigen in the lungs early post-infection. Surface staining of the viral HA revealed that most cell populations become infected, most prominently CD45neg cells, alveolar macrophages and neutrophils. Our existing data highlight the cellular heterogeneity and complexity of antigen-bearing cells within the lung and their potential as secondary points of contact for infiltrating T cells or targets for tissue resident memory cells established by previous infection.
Although it is well established that influenza A virus infection is initiated in the respiratory tract, the sequence of events and the cell types that become infected or access viral antigens remains incompletely understood. In collaboration with Dr. Luis Martinez, we have created and studied a novel Influenza A/California/04/09 (H1N1) reporter virus that stably expresses the Venus fluorescent protein. This has allowed us to identify antigen-bearing cells over time in a mouse model of infection using flow cytometry. Thus far, our studies have revealed that many hematopoietic cells, including subsets of monocytes, macrophages, dendritic cells, neutrophils and eosinophils acquire influenza antigen in the lungs early post-infection. Surface staining of the viral HA revealed that most cell populations become infected, most prominently CD45neg cells, alveolar macrophages and neutrophils. Our existing data highlight the cellular heterogeneity and complexity of antigen-bearing cells within the lung and their potential as secondary points of contact for infiltrating T cells or targets for tissue resident memory cells established by previous infection.