Circuit Tracing
Neuroanatomy is the foundation of modern neuroscience. Recent technical innovations have made it possible to reconstruct large numbers of neurons in order to gather “high throughput” information about the different morphological cell types in the brain. One of the overarching goals of my research is to link structure and function in the visual system. In other words, we believe that in order to understand the function of a particular neuron in the visual system, we must understand its morphology and how it is wired up in the network, in addition to measuring its physiological responses to visual inputs.
We have explored a number of different neuroanatomical techniques to identify the morphology and connectivity of specific neurons in the visual system. Recently we have utilized a modified rabies virus that acts like a retrograde tracer with the added benefit of enabling complete reconstruction of the full dendritic morphology of infected neurons. Rabies normally infects neurons by entering at the synapse on the axon terminal. The virus then moves in the retrograde direction to infect the cell and its dendrites where it can cross a synapse to then infect the presynaptic neurons connected to the host cell. Viral entry at the synapse is dependent upon a viral coat protein (a glycoprotein translated from the “G” gene in the viral genome). Therefore, removal of the G gene renders the virus unable to cross synapses and infect additional neurons beyond the initial infected host cell. We take advantage of the exclusively retrograde nature of rabies to label and identify neurons projecting to specific targets such as the LGN.
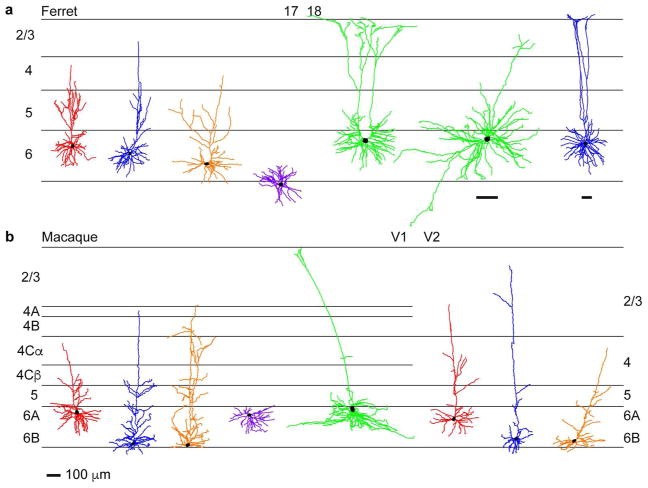
An additional benefit of using rabies virus to trace neuronal circuits is that expression is robust, making it possible to identify rare and previously undiscovered cell types. For example, we recently characterized corticogeniculate feedback circuits connecting the secondary visual cortex (V2) with the LGN (Briggs et al., 2016). Furthermore, the extensive labeling of many individual neurons with rabies virus enables statistically robust characterization of distinct morphological cell types among a large sample of reconstructed neurons (Bragg & Briggs, 2017). Taking advantage of these benefits, we discovered that neurons in the thalamic reticular nucleus (TRN) that project to and inhibit LGN relay neurons are morphologically diverse. However, their diversity does not reflect the parallel stream segregation that is present in the LGN. Instead, TRN morphological diversity may reflect specialization of acuity processing in highly visual animals (Bragg et al., 2017). Similar robust analyses also revealed that corticogeniculate neurons are diverse and separable into at least three distinct subtypes, which may underlie parallel streams of feedback mirroring the feedforward visual parallel streams (Briggs et al., 2016; Hasse et al., 2019). Thus modern circuit tracing methods coupled with sophisticated quantification techniques can provide important new insight into the structural organization and wiring of circuits in the visual system. Future work will focus on using our extensive dataset to characterize the less common subcortical circuits that contribute to visual processing, including under-studied projections from extrastriate cortex to the LGN that could contribute to residual visual perception in blindsight.
For examples of individual reconstructed neurons from Briggs et al., 2016, Bragg et al., 2017, and Hasse et al., 2019, please refer to the atlas generated at NeuroMorpho.org.