Integration of Longevity Signals by the Myc-Family of Transcription Factors
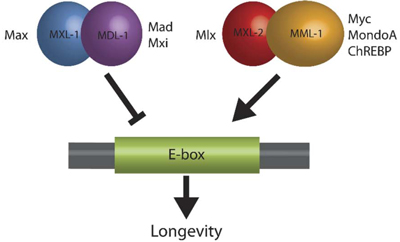
Figure 1. The C. elegans Myc-Mondo/Mad complexes have opposing functions in longevity (from Johnson et al. 2014 PLoS Genetics)
We discovered a vital role for the Myc family of transcription factors in regulating transcriptional programs that set the progression of aging in C. elegans. Myc and the related Myc family members have well known roles in diverse biological contexts, but our discovery directly links the Myc family to aging. In C. elegans two heterodimeric complexes have opposing roles in aging and transcriptional control (homologous to mammalian Myc-Mondo and Mad complexes) (Figure 1). These complexes interact genetically and molecularly with both insulin/IGF signaling and with dietary restriction. Thus, the Myc family of transcription factors represents a newly discovered convergence point for these central aging related pathways (Figure 2).
The Myc-Mondo and Mad transcriptional complexes represent an evolutionarily conserved entity for coupling nutrient sensing, metabolism, and aging. ChREBP, one of two mammalian Mondo homologs, is one of 26 genes lost in Williams-Beuren Syndrome (WBS), which among other symptoms causes metabolic dysfunctions, silent diabetes, and premature aging. Characterizing the role of Myc transcription factors may facilitate the development of rationale strategies for the treatment of age-associated disease and metabolic disorders (e.g. diabetes, glucose intolerance, and insulin resistance) to promote human health.
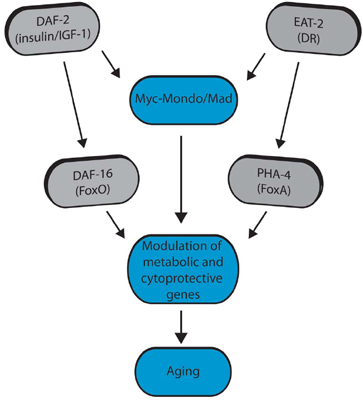
Figure 2. Model for Myc-Mondo/Mad transcription factors in longevity control (from Johnson et al. 2014 PLoS Genetics).
Recently we have focused on dissecting the role of the Myc-family in conferring the benefits of dietary restriction (DR). Understanding these molecular mechanisms are important as DR delays the onset of aging-associated diseases including sarcopenia, metabolic syndrome conditions, and neurodegeneration. We have identified thousands of differentially expressed genes under conditions of DR, which are dependent on mxl-2 (ortholog of mammalian Mlx). Functional enrichment of components of the mxl-2-dependent components of the DR transcriptional signature implicate an essential role for mxl-2 in metabolic shifts initiated by chronic DR. Additionally, many germ line and gonadal genes decrease expression, implying that the Myc family members are crucial for reallocation of resources between the germline and soma that is initiated by DR. Elucidating the molecular mechanisms through which the Myc-family regulates the adaptive transcriptional response to DR is a continued area of active investigation.