Our Research
Project 1: What is the molecular basis regulating cbDVG generation in RSV?
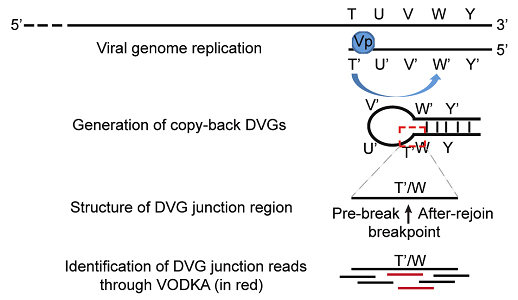
Schematic representation of cbDVG generation from negative strand viruses and general principle behind VODKA identification of cbDVGs. The red dashed square marks the unique junction region that distinguishes DVGs from full-length viral genomes. This junction region was used by VODKA to identify cbDVGs.
During cbDVG generation, the viral polymerase detaches from the viral template at a “break point”, T, and resumes elongation towards the 5’ end of the nascent strand at a downstream “rejoin point”, W’, to form the unique junction region (Figure 2). Previously, it was thought that the locations of break and rejoin points were totally random. By combining RNA-seq with Viral Open-source DVG Key Algorithm (VODKA, algorithm detecting DVGs), we found that rejoin points concentrate primarily in three genomic hotspots, whereas the break hotspots are more widely distributed (Sun et al, 2018). Importantly, specific nucleotide substitutions within rejoin hotspot dramatically change the pattern where cbDVGs were formed. We hypothesize that these genomic hotspots may contain important signals to control cbDVG generation. In this project, we aim to identify and investigate these signals regulating cbDVG rejoin points and break points, as well as the mechanisms by which viral polymerase recognizes and interprets those signals.
For more info see publication (Sun et al, 2018).
Project 2: Are there common rules controlling cbDVG generation among different viral families?
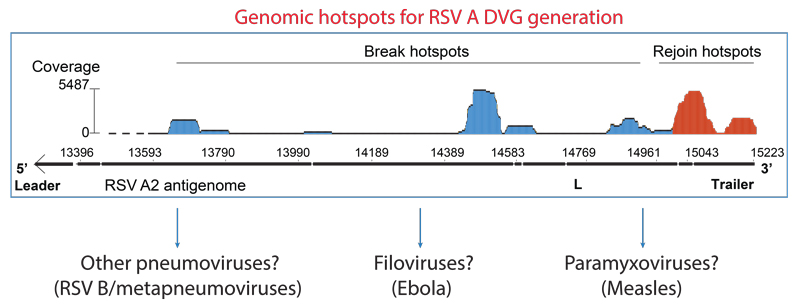
Are cbDVG generation of other viruses regulated similarly as RSV?
cbDVGs are generated during infection of many single-stranded negative-sense viruses, including paramyxoviruses (measles and parainfluenza), pneumoviruses (metapeumoviruses and RSV), and filoviruses (Ebola). These cbDVGs are all reported to have immunostimulatory functions in vitro and are associated with persistent infection in infected cell cultures. What are the mechanisms of cbDVGs formation in other negative-sense RNA viruses beyond RSV? Since all cbDVGs contain break and rejoin points, is it possible that cbDVG generation shares similar mechanism regardless of virus families? In this project, we will aim to answer this intriguing question. We will create a similar minigenome system used in RSV to measles virus and Ebola virus. The ultimate goal of this project is to investigate the mechanism of cbDVG generation in other negative-sense RNA viral families, which can be utilized to control de novo cbDVGs across virus families.
Project 3: Can we manipulate de novo cbDVG generation during viral replication?
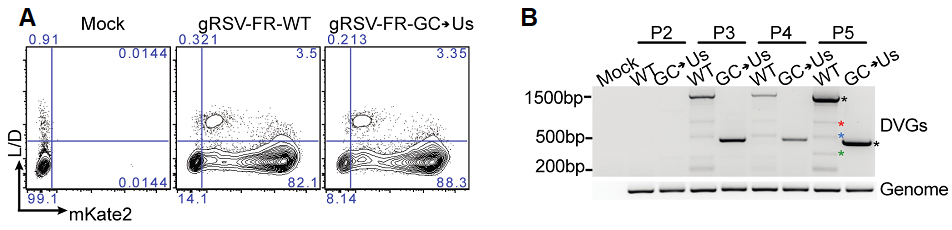
A conserved rejoin region determines cbDVG formation during viral infection in vitro. (A) Hep2 cells were infected with gRSV-FR-WT-P3 and gRSV-FR-G-C->Us-P3 at MOI of 1.5 and harvested at 72h post infection and stained for aqua L/D, followed by flow cytometry for mKate2 expression as a proxy for virus replication. (B) cbDVG dedetection from P2, P3, P4, and P5 of gRSV-FR-WT and gRSV-FR-GC>Us via DVG specific RT-PCR using DI1/DI-R primer set in HEp2 cell. Confirmed cbDVG-like fragments are labeled by asterisks next to the gel.
As DVGs can impact viral pathogenesis through multiple ways (Figure 1), modulating DVG generation will be a novel way to understand and influence viral pathogenesis. Due to the lack of knowledge of DVG generation, it is thought that manipulating the ability of a virus to generate DVGs, or de novo DVG generation, is impossible. Our findings suggest that this process, although with certain level of variations, is regulated by viruses and possibly hosts. Recently, we have reduced the GC contents in one of the identified rejoin hotspots and found that this mutation reduced the number of cbDVG species generated during RSV infection significantly (Figure 3), further proving the feasibility to manipulate de novo cbDVG generation. This project aims to generate mutant viruses to modulate the species, amount, and kinetics of cbDVG generation during infection via RSV reverse genetic system. Using these viruses, we will study how these aspects of cbDVGs influence host responses, viral pathogenesis and viral evolution.
Project 4: The generation and function of DVGs in SARS-COV-2
Coronaviruses (CoV) are a family of single-stranded positive-sense. This family includes several contagious human pathogens, such as SARS-CoV, MERS-CoV, and SARS-CoV-2. CoV replication generates DVGs that contain deletions of genomic sequences but retain 5’ and 3’ genomic untranslated regions and thus those DVGs belong to another type, named deletion DVGs. However, the function and generation of CoV deletion DVGs are not well understood. In this project, we will focus specifically on SARS-CoV-2 aiming to address the following questions:
- What are the host responses induced by DVGs during SARS-CoV-2 infection?
- Are DVGs present in COVID-19 patients? If so, what are their functions and does their presence impact clinical outcomes?
- What are the viral determinants for CoV DVG generation? Can the pathogenesis of SARS-CoV-2 be altered by manipulating the generation of DVGs?