Welcome to the Yao Lab
Regulatory non-coding RNAs and RNA-binding proteins (RBPs) are important research areas in gene regulation and RNA biology. The Yao Lab’s research includes two themes: 1) Determining the pathophysiological function and regulatory mechanisms of non-coding RNAs and RNA-binding proteins in the cardiac system and in cardiovascular disorders; 2) Elucidating the role of translation machinery and translational control in the cardiovascular system and identifying therapeutic targets for the treatment of cardiac diseases. The overall focus of the Yao Laboratory is to elucidate the pathophysiological function and molecular mechanism of translational control in cardiac health, disease, and crosstalk to other cardiac physiological processes.
The Yao Lab has recently discovered a new type of stress-responsive, protein-directed human RNA switch that regulates expression of vascular endothelial growth factor-A in human monocytic cells (Ray, PS, et al. Nature 2008; Yao, P, et al. Plos Biology 2013) that may play a role in cardiovascular disorders and cancers. We also identified a novel mRNA processing mechanism that expands the human proteome at the posttranscriptional level and regulates gene expression (Yao, P, et al. Cell 2012).
Dr. Yao is currently asking four major scientific questions in his research:
- What is the impact of translational control in heart physiology and pathology?
- What are the molecular mechanisms of translational regulation in cardiac biology?
- How does translation control connect to other biological processes (e.g., transcriptional control, mitochondrial biology, etc.) in the heart?
- What is the implication of translational regulation in heart disease treatment?
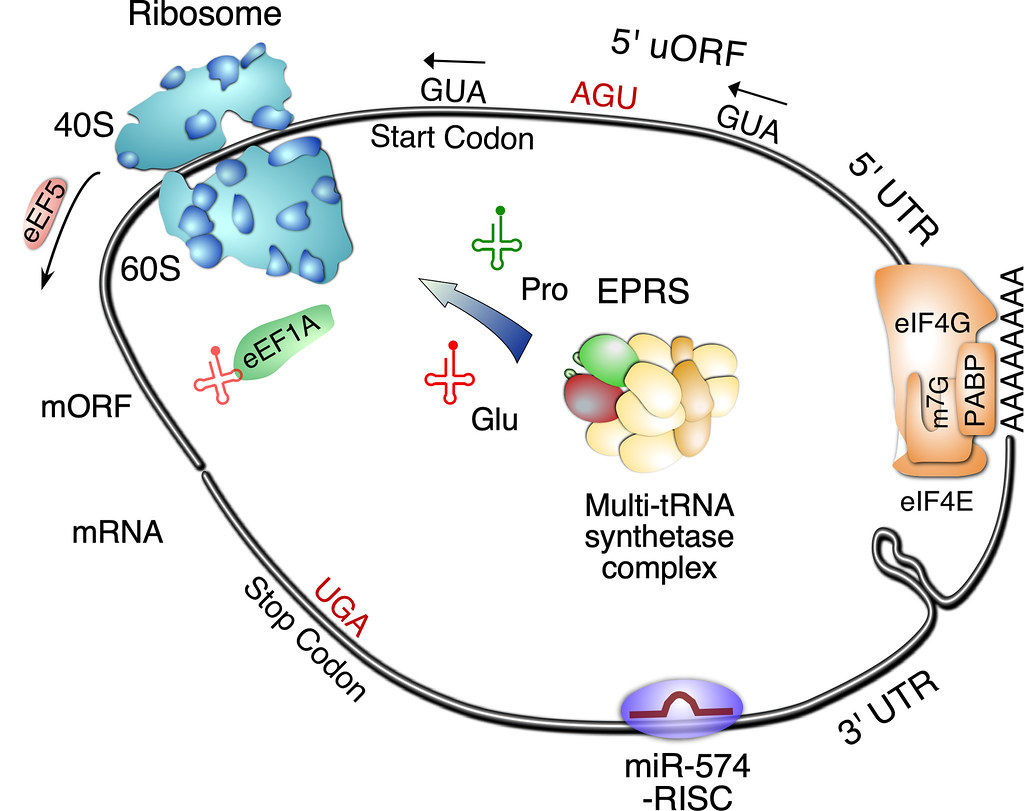
The Yao Lab recently discovered the molecular mechanism of human glutamyl-prolyl-tRNA synthetase (EPRS) directed translational regulation of proline-rich extracellular matrix protein during cardiac fibrosis and the action of EPRS-targeting drug halofuginone in inhibiting pro-fibrotic protein synthesis at the genome-wide level (Circulation Research, 2020). In this work, we demonstrate that multiple cardiac stresses induce the expression of glutamyl-prolyl-tRNA synthetase (EPRS) that promotes cardiac fibrosis via increased Pro-tRNAPro pool and consequent translation of pro-fibrotic proline codon rich mRNAs (e.g., collagens, LTBP2, SULF1 among others) in cardiac fibroblasts using genome-wide polysome-seq and RNA-seq. This project will provide new insights into the translational control mechanisms in cardiac fibrosis and identify novel disease markers and therapeutic targets to treat heart disease.
We reported the role of miR-574 in regulating VEGFA mRNA translation, which involves RNA switches during the interplay between microRNA and RNA-binding protein based on our recent findings (Nucleic Acids Research, 2017; BBRC, 2019; CSBJ, 2019). We also elucidated the role of the miR-574-FAM210A axis and FAM210A in regulating mitochondrial translational homeostasis and cardiac remodeling (EMBO Molecular Medicine, 2021; Cardiovascular Research, 2023 in press). In addition, we discovered the role of miR-574 host gene FAM114A1 in regulating Ang II signaling and cardiac remodeling (JCI Insight, 2022). Importantly, we found that congenital heart disease and cancer associated protein PRRC2B functions as a novel RNA-binding protein and regulates translation initiation of cell cycle progression related mRNAs, thereby promoting cell proliferation and possibly influencing organ development and cancer formation (Nucleic Acids Research, 2023). This is a missing piece of the “Yamanaka translation factors” hypothesis that is essential for full understanding of embryonic stem cell proliferation or differentiation. In a most recent manuscript, we discover that dsRNA structure regulates "see-saw" translation balance of uORF & mORF and can be targeted by potential ASO RNA drugs (https://doi.org/10.1101/2023.06.15.545153).
We employ various approaches and methods of biochemistry and biophysics, molecular and cellular biology, genetic and surgical mouse models, high throughput multi-omics, and bioinformatic analytic tools to dissect the convergent and divergent regulatory pathways. The long-term objective of our laboratory is to identify novel translation-related molecular mechanisms that control gene expression and conduct pathophysiological function in cardiac system, as well as to develop novel therapeutic approaches for the prevention or treatment of human cardiovascular diseases. We currently have NIH funding to support a research program of translational control of human health and disease and industry funding from Novo Nordisk to develop novel RNA-based drug for treating fibrosis related human diseases including heart failure, liver stenosis, and cancer.
We are actively recruiting postdoctoral associate, staff scientist, and graduate student positions. Please see details in the other Yao Lab webpage and in the Yao Lab’s Twitter: @YaoLabURMC.

Peng Yao, Ph.D.
Principal Investigator
Publications
View All Publications- Translational Control in Cardiac Pathophysiology and Therapeutic Development: When mRNA Meets the Heart.; International journal of molecular sciences; Vol 26(16). 2025 Aug 14.
- Upstream open reading frame inactivation augments GATA4 translation and cardiomyocyte hypertrophy in mice.; bioRxiv : the preprint server for biology. 2025 May 18.
- Newborn apical resection preserves the proliferative capacity of cardiomyocytes located throughout the left ventricle.; Stem cells (Dayton, Ohio). 2025 Apr 15.
- FAK and p130Cas modulate stiffness-mediated early transcription and cellular metabolism.; bioRxiv : the preprint server for biology. 2024 Jan 16.
- Cardiomyocyte-Specific Loss of Glutamyl-prolyl-tRNA Synthetase Leads to Disturbed Protein Homeostasis and Dilated Cardiomyopathy.; Cells; Vol 13(1). 2023 Dec 22.
- Cardiomyocyte-specific Loss of Glutamyl-prolyl-tRNA Synthetase Leads to Disturbed Protein Homeostasis and Dilated Cardiomyopathy.; bioRxiv : the preprint server for biology. 2023 Sep 22.
Affiliations
- Medicine
- Biochemistry & Biophysics
- Aab Cardiovascular Research Institute
- Center for Biomedical Informatics
- Center for RNA Biology: From Genome to Therapeutics
- NIH T32 Training Grant in Cellular, Biochemical & Molecular Sciences
- Biochemistry & Molecular Biology Ph.D. Program
- Biomedical Genetics and Genomics Ph.D. Program
- Cellular and Molecular Pharmacology and Physiology Ph.D. Program
- PhD Program in Pathology - Cell Biology of Disease
- Mitochondrial and Metabolic Research Center
Lab Photos
Contact Us
Yao Lab
601 Elmwood Ave
Box CVRI
Rochester, NY 14642