CGRP Signal Transduction
Our lab is studying the neuropeptide calcitonin gene-related peptide (CGRP), focusing on the role of CGRP in physiology and pathophysiology, and on the biochemical mechanism of signal transduction at the CGRP receptor. To facilitate these studies we are collaborating to develop new methods for high efficiency gene transfer into mammalian cells using arrays of carbon nanotubes. Work in the lab combines approaches from molecular biology, biochemistry, cell biology, and engineering.
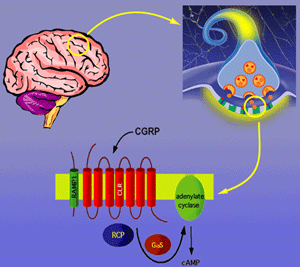
CGRP receptor complex: Trimer of CLR, RAMP1 and RCP required for generation of cAMP signaling.
CGRP is a 37 aa peptide with potent vasodilator activity, that has been studied for its role in regulating vascular tone. It has also been shown to be important for migraine, regulating immune function, and perception of pain. Despite these important physiologic roles, therapeutic strategies targeting CGRP and its receptor have been limited. This delay has primarily been due to the fact that CGRP is a peptide, and thus has a complicated biosynthesis with a short half-life, and also due to the fact that the receptor for CGRP has turned out to be a complex of proteins, rather than a single molecule as predicted.
CGRP Receptor
The complexity of the receptor for CGRP is particularly important. One of the central questions in biology today is how a cell converts an extracellular signal to an intracellular chemical signal that the cell can interpret. For example, extracellular hormones by themselves have no meaning to a cell until they bind their cognate cellular receptor and stimulate an intracellular signal. This process, known as signal transduction, involves a change in the three-dimensional structure of the receptor, which results in activation of intracellular signaling mechanisms. Such intracellular signals are known as second messengers, and can take the form of chemicals produced in response to receptor activation such as cyclic adenosine monophosphate (cAMP), cGMP, inositol tris phosphate (IP3), or as altered ion concentrations (sodium, potassium, calcium, chloride) as a result of ion channel activation, or as activation of kinases or phosphatases, or changes in gene expression.
It was once thought that G protein-coupled receptors (GPCRs) functioned as monomers in the cell membrane. However, recent data support the model that many GCPRs require homo- and heterodimerization for function. We discovered an intracellular protein required for activation of the GCPR for CGRP. The receptor for CGRP is an attractive target for development of therapeutics given the wide range of physiologic roles ascribed to CGRP. However, the CGRP receptor was difficult to identify, and cloning and characterization was the result of work in multiple labs. We now know that one of the reasons the CGRP receptor was so difficult to identify is that it is composed of three proteins: (1) A ligand-binding receptor named calcitonin-like receptor (CLR) that has the stereotype GCPR heptahelical transmembrane conformation (2) An accessory protein named receptor activity modifying protein (RAMP1) that acts as a molecular chaperone and also contributes pharmacologic specificity (3) A second accessory protein named receptor component protein (RCP) that couples the CLR/RAMP1 complex to the cellular signaling machinery.
RCP is a novel regulator of GPCR function, and our laboratory is working to discover the mechanism of RCP function and the extended role of RCP in cellular physiology. RCP interacts directly with CLR to confer activity, and this interaction is observed in cell culture and in vivo, where RCP-CLR interaction increases the efficacy of CGRP. Areas of focus are (a) determining the molecular and biochemical requirements for RCP (b) Discovering proteins that interact with RCP in a functional receptor complex using proteomic strategies (c) Determining the role of RCP and CGRP in vivo using targeted homologous recombination.
Carbon Nanotube-Mediated Gene Transfer
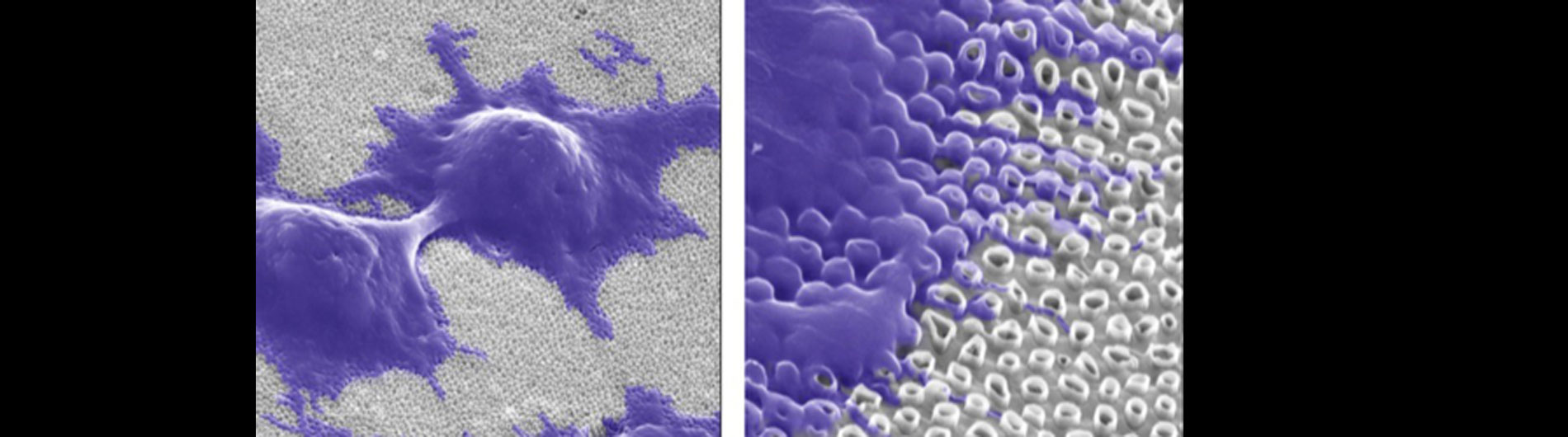
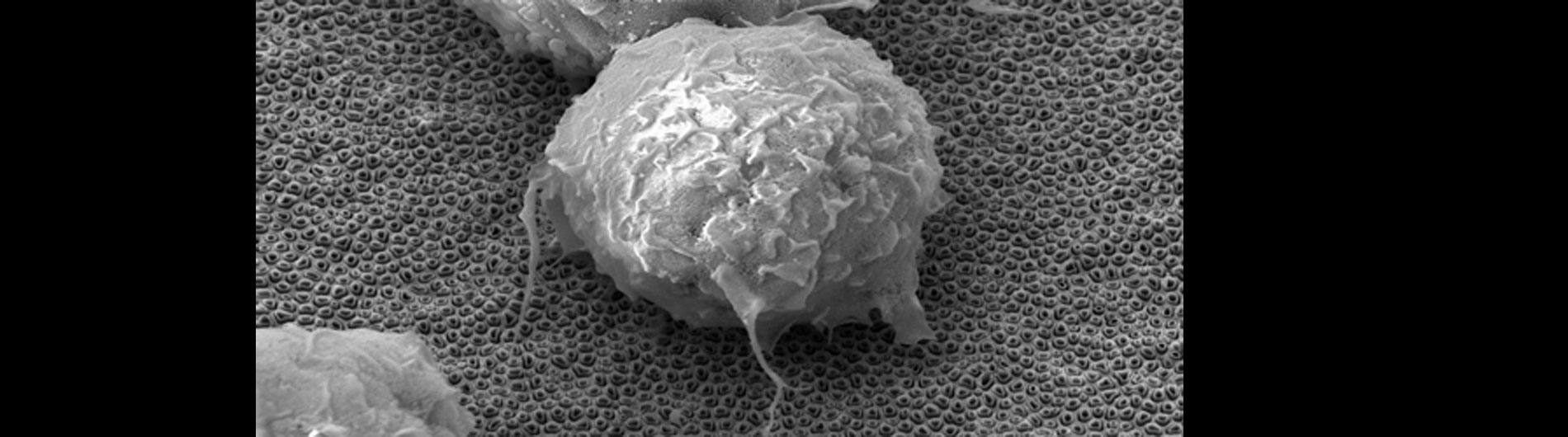
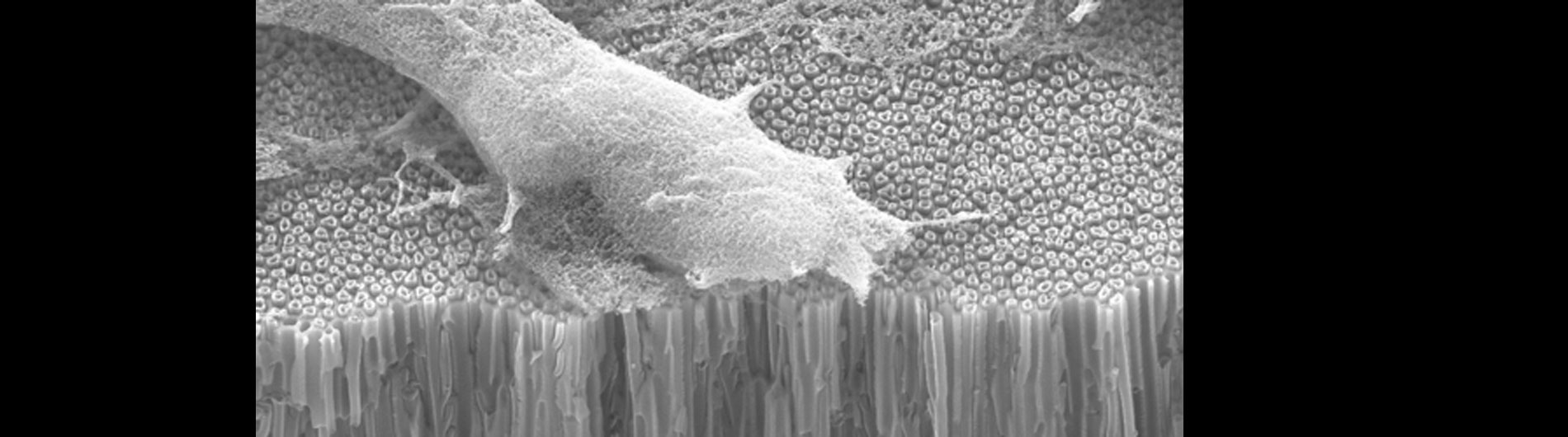
Our lab is also working on developing novel technologies to transfer nucleic acids and proteins into cells using nanoscale devices, for enhanced gene editing (CRISPR/Cas9), production of induced pluripotent stem cells (iPSC), and chimeric antigen receptor in T-cell (CAR-T) cancer therapy. Current gene transfer technology, including lipofection, electroporation, and viral delivery, has enabled break-through advances in basic and translational science. Despite this success there are limitations to current gene transfer technology: many cell types cannot be consistently transfected by lipofection or electroporation (stem cells, iPSCs, T-cells) and viral delivery has limitations to the size of experimental DNA that can be packaged.

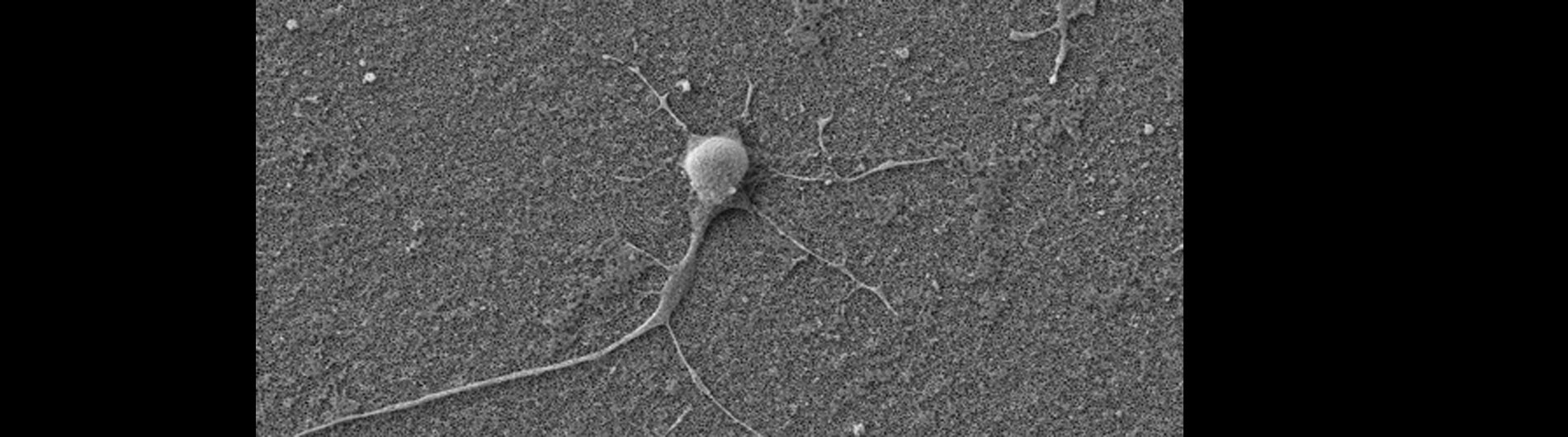
Human primary neuron growing on carbon nanotube array.
In collaboration with the Schrlau lab (Dept. Mechanical Engineering, RIT), we have developed a coverslip-like device consisting of millions of 200 nm-diameter hollow carbon nanotubes (CNT) embedded in a base that has enhanced gene transfer capabilities, including an expanded range of target cells, high efficiency, low toxicity, and the potential to transfer mixed combinations of protein and nucleic acids. Manufacture of these CNT devices is readily scalable, and we have developed a simple protocol to use these CNT devices for high efficiency gene transfer. We are working to optimize our carbon nanotube devices for gene transfer into difficult to transfect cells such as stem cells, iPSC, and primary T-cells, to facilitate enhanced biochemical analysis of cells, CRISPR gene editing, and production of cells for CAR-T anti-cancer therapy.

Ian M. Dickerson, Ph.D.
Principal Investigator
Publications
View All Publications- Loss of Calcitonin Gene Related Receptor component protein (RCP) in nervous system can bias "gepant" antagonism.; bioRxiv : the preprint server for biology. 2024 Oct 26.
- Uptake of severe acute respiratory syndrome coronavirus 2 spike protein mediated by angiotensin converting enzyme 2 and ganglioside in human cerebrovascular cells.; Frontiers in neuroscience; Vol 17, pp. 1117845. 2023 Feb 16.
- Calcitonin gene-related peptide: An intra-articular therapeutic target for TMJ disorders.; Clinical and experimental dental research. 2022 Jun 14.
- Inhibition of CGRP signaling impairs fracture healing in mice.; Journal of orthopaedic research : official publication of the Orthopaedic Research Society. 2022 Jan 24.
News
Affiliations
- Neuroscience
- Biomedical Engineering
- Pharmacology & Physiology
- Del Monte Institute for Neuroscience
- Intellectual & Developmental Disabilities Research
- Biochemistry & Molecular Biology Ph.D. Program
- Cellular and Molecular Pharmacology and Physiology Ph.D. Program
- Neurobiology & Anatomy Ph.D. Program
- Neuroscience Ph.D. Program
Contact Us
Dickerson Lab
MC 6-8562
601 Elmwood Ave,
Rochester, NY 14642
(585) 276-5334