Regulation of Gene Expression by Targeted Degradation of RNA
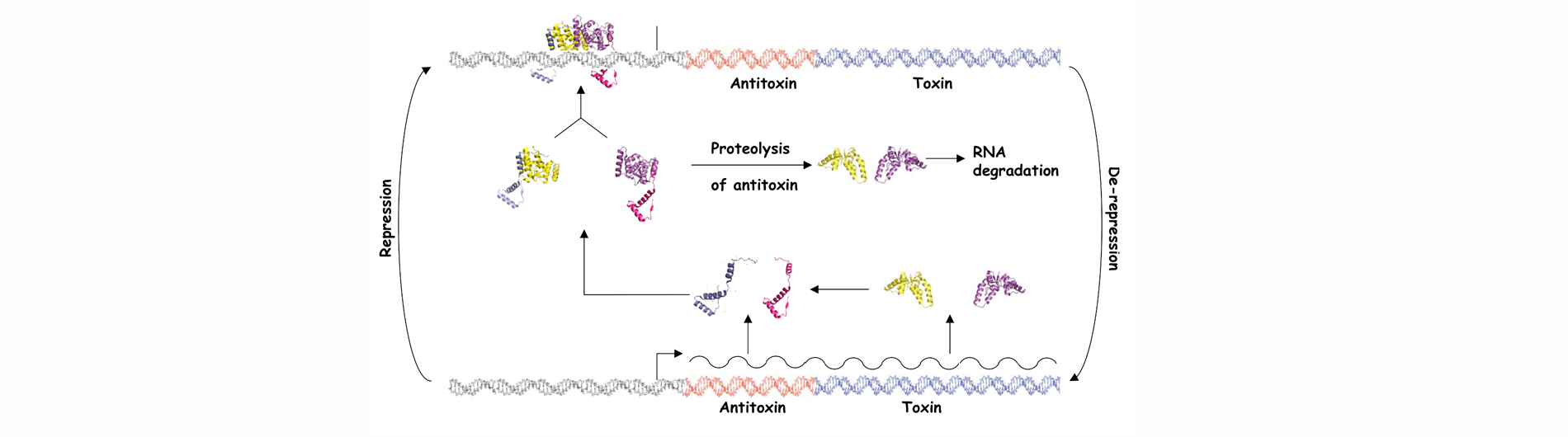
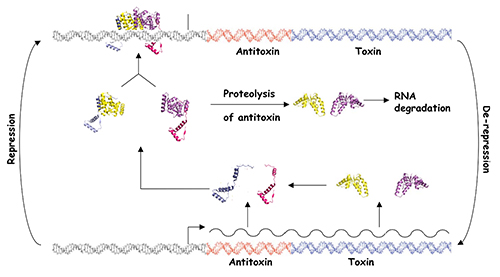
Bacteria have evolved complex systems to facilitate their survival under conditions of stress such as temperature fluctuations, oxidative damage and nutrient deprivation. Stress often causes pervasive gene regulatory responses that minimize metabolic activity and result in a dormant state. Dormant bacteria are largely resistant to antimicrobial therapeutics, which complicates treatment of latent bacterial infections. Moreover, it appears that stochastic processes elicit the formation of small numbers of persister cells within growing populations of bacteria. These metabolically dormant cells are tolerant to drugs that kill replicating cells, which may result in the resurgence of infection after cessation of therapy. Bacterial toxin-antitoxin systems represent one adaptation that down-regulates cellular metabolism in response to stress and contributes to the formation of dormant cells and persisters. Type II TA systems feature protein antitoxins that bind to and inactivate protein toxins inside bacterial cells. Cells subjected to certain types of stress, such as nutrient deprivation, produce proteases that cleave the antitoxin, thereby releasing the active toxin. Many of the known Type II toxins inhibit protein synthesis, which leads to reversible dormancy. We use genetic and molecular biology techniques to study the RNA targets of VapC toxins and the specificity determinants that govern VapB-VapC TA interactions. We have also developed a generalizable, whole cell, high throughput screening assay for the identification of small molecule inhibitors of TA interactions from any bacterium. The compounds identified with this assay will provide unique tools for the study of TA systems in bacteria.
A second area of the work in the laboratory focuses on understanding the mechanisms that ensure the proper expression of messenger RNAs in eukaryotes. The research focuses on a nuclear mRNA surveillance pathway in eukaryotes that destroys aberrant RNAs before they exit the nucleus. Central to this pathway is a conserved complex of proteins called the RNA exosome, which degrades RNA molecules that fail to undergo post-transcriptional processing reactions such as polyadenylation and splicing. A major unanswered question addressed by the research is what components of the nuclear exosome and the associated TRAMP complex are required for the processing and degradation of RNAs in the nucleus. This RNA surveillance system guards against the formation of defective RNA-protein complexes that have toxic effects on cell growth and proliferation. Previous studies in our group identified Rrp6p, a nuclear exoribonuclease component of the exosome and showed that it plays a critical role in degrading aberrant RNAs in the nucleus. More recent evidence indicates that a second complex called TRAMP plays a key role in activating RNA substrates for degradation by the nuclear exosome. Experiments underway in the lab aim to (i) identify the components of the TRAMP complex required for enhancement of Rrp6p activity, (ii) elucidate the role that TRAMP and Rrp6p play in a general and a specific pathway for mRNA degradation.

James S Butler, Ph.D.
Principal Investigator
Publications
View All Publications- Toxins targeting transfer RNAs: Translation inhibition by bacterial toxin-antitoxin systems.; Wiley interdisciplinary reviews. RNA. 2018 Sep 16.
- Homologous VapC toxins inhibit translation and cell growth by sequence specific cleavage of tRNA(fMet).; Journal of bacteriology. 2017 Nov 06.
- Structural determinants for antitoxin identity and insulation of cross talk between homologous toxin-antitoxin systems.; Journal of bacteriology. 2016 Sep 26.
- Using S. cerevisiae as a Model System to Investigate V. cholerae VopX-Host Cell Protein Interactions and Phenotypes.; Toxins; Vol 7(10). 2015 Oct.
Contact Us
Butler Lab
MRBX 2-11211
601 Elmwood Ave
Rochester, NY 14642