The Roles of the Genetic Code in Translation
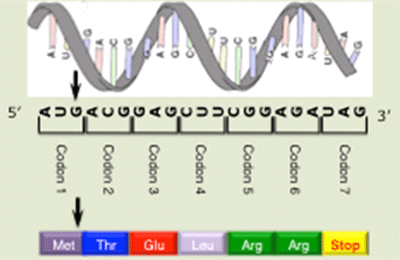
The genetic code and protein synthesis
We study the role of the genetic code in controlling the rate and efficiency of translation in the eukaryote Saccharomyces cerevisiae. Translation of the genetic code into functional proteins is a fundamental process, mediated by concerted actions of the ribosome and amino-acylated tRNAs interacting with codons in the mRNA. Translation influences both the composition and identity of the proteome, as translation of different genes occurs with remarkable differences in efficiency and is regulated both by global changes in cellular conditions as well as by the genetic code itself. The genetic code includes 61 triplet codons that specify insertion of the twenty amino acids into proteins resulting in many synonymous codons that specify insertion of the same amino acid. The choice of synonymous codons used to encode a protein affects the efficiency of translation, the accuracy of amino acid insertion, protein folding, and reading frame maintenance. However, neither the mechanism nor the biological significance of codon-mediated regulation is understood, particularly in eukaryotes.
One focus is to deduce the magnitude as well as the mechanisms of codon-mediated effects by identifying codons and codon combinations that are responsible for reduced translational efficiency and by defining the parameters that modulate codon-mediated regulation. To this end, we performed the first systematic analysis of codon effects on expression. We discovered that decoding interactions at neighboring sites within the ribosome play an important role in mediating codon effects on translation efficiency, particularly for arginine CGA codons. We are currently screening libraries of codon combinations using fluorescent reporters to identify other inhibitory codon combinations and define the common features of such combinations. Moreover, analysis of the use of inhibitory codon combinations in the yeast genome should provide initial insight into the biological role of these codons.
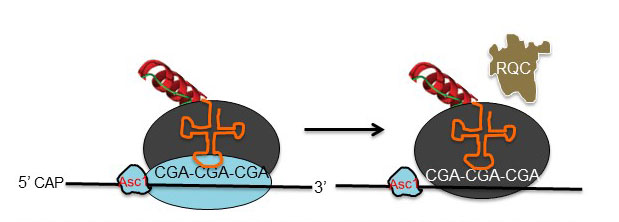
Model of translation through internal CGA codon repeats. The ribosome stalls, continued translation is blocked by ribosomal protein Asc1, ribosomal subunits are split, and the nascent polypeptide is targeted for degradation by the ribosomal quality control complex (RQC)
A second focus is to deduce the mechanism(s) by which inhibitory codons exert their effects on the elongating ribosome. Both biochemical and genetic approaches are being used to study the basis for the inhibitory effects caused by decoding arginine CGA codon pairs. We have determined that ribosomes recruit the E3 ubiquitin ligase Ltn1, a part of the ribosome quality control complex, at CGA codon repeats, thus targeting the nascent polypeptide for degradation. Structures of the yeast eukaryote ribosome and of the ribosome quality control complex provide a framework for understanding the mechanisms by which factors identified in the genetic screen exert their effects.