URMC / Labs / Ermolenko Lab / Projects / Structural rearrangements of the ribosome and translation factors
Structural rearrangements of the ribosome and translation factors
Protein synthesis by the ribosome is accompanied by a number of large conformational changes. For example, the ribosome undergoes cyclic forward and reverse rotations between the large and small ribosomal subunits. Conformational changes within ribosomes are coupled to structural rearrangements of translational factors that bind to the ribosome to mediate different steps of protein synthesis. We use singe-molecule FRET to establish the intricate sequence of these structural rearrangements in ribosomes and translation factors (https://www.ncbi.nlm.nih.gov/pubmed/25288752 , https://www.ncbi.nlm.nih.gov/pubmed/24324137 , https://www.ncbi.nlm.nih.gov/pubmed/25463439, https://www.ncbi.nlm.nih.gov/pubmed/26668356 ).
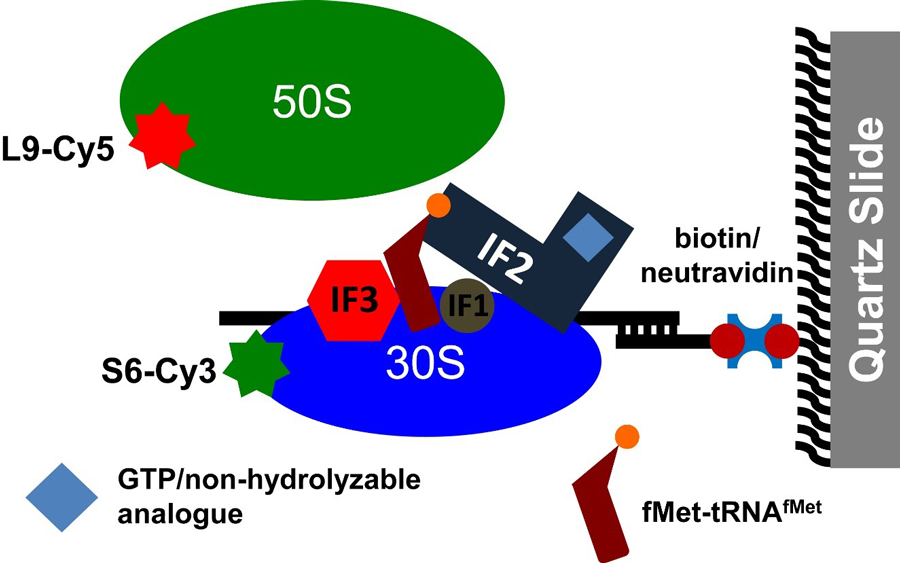
Figure legend: Following intersubunit rotation during subunit joining by FRET. Cy3-labeled 30S initiation complexes (IC) formed in the presence of mRNA, fMet-tRNAfMet, IF1, IF2, IF3 and GTP (or GDPCP) were immobilized to a quartz slide by NeutrAvidin and a biotinylated DNA primer annealed to the mRNA. Cy5-labeled 50S subunits were delivered to the 30S ICs during imaging and subunit joining was detected by the appearance of FRET between Cy3 and Cy5. (From https://www.ncbi.nlm.nih.gov/pubmed/26668356 ).
« back to all projects