Welcome to Microbiology & Immunology

Chair, Tanya Mayadas, PhD shares vision for the future
My vision for the department builds on the strong foundation previously established by my colleagues and focuses on five key priorities: recruiting and supporting outstanding researchers; fostering collaborative translational research across disciplines; expanding grant and philanthropic support to grow our portfolio and infrastructure; strengthening integration with the Institute of Immunological Sciences (IIS) to amplify immunology-focused research; and enhancing our teaching curriculum to promote cross-training and prepare the next generation of scientists and clinicians. I look forward to advancing these goals and strengthen our impact together.
Department Highlights
- The department is home to two NIH-funded training grants, Infection and Immunity: The Pathogenesis of Host-Microbe Interactions (T32AI118689) and Predoctoral Training Program in Immunology (T32AI007285).
- One of our graduate students was the co-inventor of the technology behind the current HPV vaccine.
- Our past students currently occupy leadership positions in academia, the biotech industry and the government.
2026 Summer Fellowship in Immunological Sciences Research
Applicants must be a University of Rochester rising sophomore or junior undergraduate with a cumulative GPA of 3.00 or greater.
Deadline: Students must apply by Friday, March 27, 2026 11:59 p.m. to be considered for this opportunity.

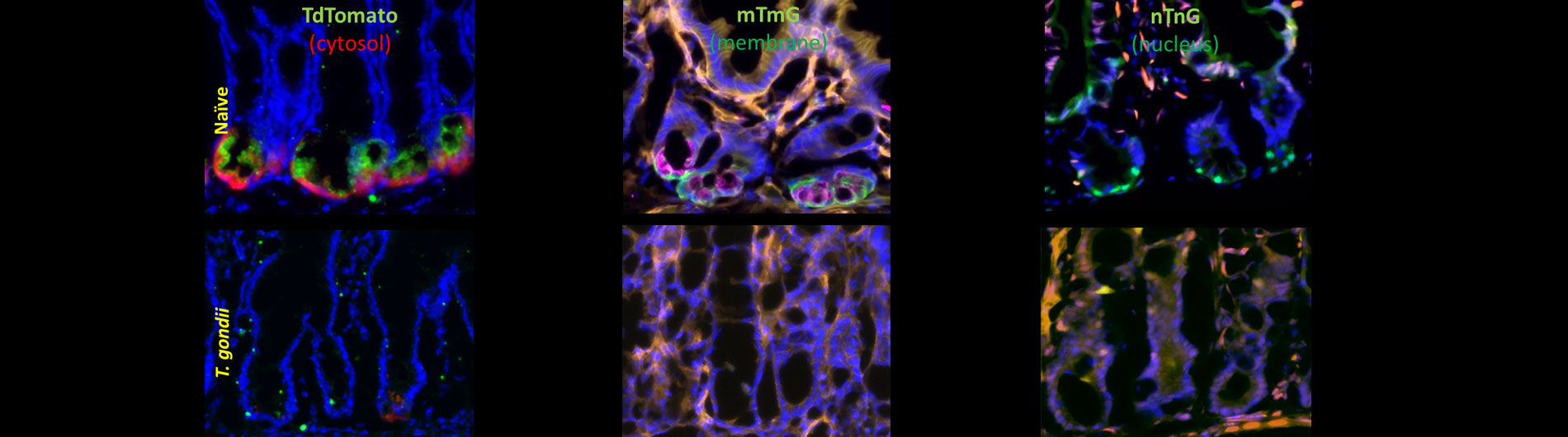
Research
Our diverse faculty's cutting edge research spans several areas of microbiology and immunology

Education
Our multiple programs prepare individuals for a career specifically focused on research and education in microbiology, immunology and virology

People
The Department of Microbiology & Immunology fosters collaborative research between laboratories and investigators.
Upcoming Events
MBI 501 Student Seminar: "Investigating Molecular Determinants that Regulate the MV-EV Transition During Orthopoxvirus Infection"
Riley Steel
Graduate Student
Thu, Mar 05 @ 12:00 PM
MC | K-307 (3-6408)