Service Analysis
The GRC performs a variety of services that generates millions of NGS data points everyday. The Bioinformatics group has worked hard to develop and maintain standard processing pipelines to reproducibly analyze bulk RNA-Seq, single cell RNA-Seq, ChIP/ATAC-Seq, and WGS/WES. These pipelines generate results that we refer to as our 'preliminary analysis' that get delivered to the investigator upon the completion of an NGS experiment.
Our deliveries are delivered directly to smdnas02 drives for URMC PIs or sent over email and provide three links to download:
(1) raw FASTQ data
(2) alignment data in BAM/BAI format
(3) multiQC report showing QC and general statistics
(4) analysis results, which will change depending on the experiment.
Below you will see all service analyses that we provide and more details about the specific results that are delivered for each.
Reference Genomes
We use all current reference genome builds and annotation versions downloaded from GENCODE. If you want your data processed with a specific genome build/version please let us know prior to the submission of your experiment.
Current human: GRCh38
Current mouse: GRCm39
Older versions can be used upon request during sample submission.
Bulk RNA-Seq
Sequencing design= Differential Expression: 1x75; Differential Isoform: 2x100
Sequencing platform = NextSeq2000/NovaSeq6000/NovaSeq X
Standard Analysis Package Includes:
(1) Aligned data files (bam files)
(2) Raw data files (fastq files)
(3) MultiQC HTML Report
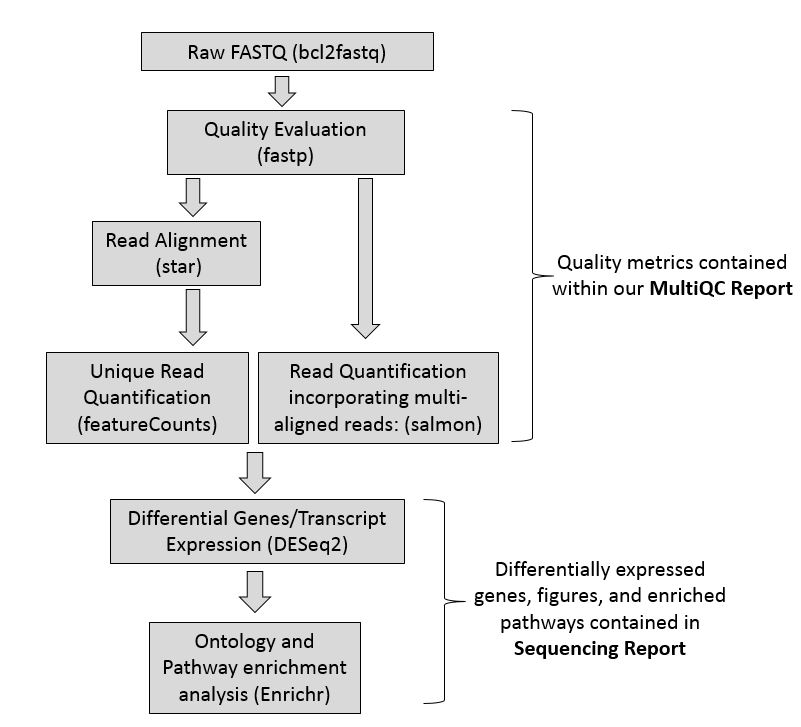
(4) Two HTML sequencing Reports (a] StarFeature Counts (Gene level quantification) and b] Salmon (Transcript level quantification))
- PCA Plot
- Sample Distance Heatmap
- Differential Expression Results for compared groups (e.g Mutant vs WT)
-- Differential Expression Summary: Basemean, log2Fold Change, Stat, p-value, p-adjusted value
-- Volcano Plot
-- MA Plot
-- Enrichr results (StarFeature Counts only)
The current software that we use to generate our preliminary results: fastp, star, multiqc, featureCounts, salmon, DESeq2, enrichr
Figure 1. Overview of the RNA-Seq Pipeline, indicating what tools are used at each step.
Download Example RNA-Seq Reports:
Please note these reports are saved within the PDF format, when we deliver the files they will be in a more interactive HTML format.
Sequencing_Report_Example MultiQC_Report_Example
The publicly available data contained within the report was downloaded from the Sequence Read Archive using accession SRP055478 and analyzed using the GRC's Bioinformatics RNA-Seq pipeline. To learn more about this project please refer to GEO accession GSE66264 and the associated publication:
Guirguis AA, Slape CI, Failla LM, Saw J et al. PUMA promotes apoptosis of hematopoietic progenitors driving leukemic progression in a mouse model of myelodysplasia. Cell Death Differ 2016 Jun;23(6):1049-59. PMID: 26742432
Click here for our Protocols.io page for more information on Bulk RNAseq analysis at the GRC.
Single Cell RNA-Seq
Sequencing paradigm = Custom Paired-End sequencing
Sequencing platform = target 50-100k reads/cell (# cells captured may change platform selection)
The GRC has the capability of running single cell experiments using 10x Chromium or a FACS plate based technique. Primarily all of the single cell experiments at URMC are executed taking advantage of the Chromium platform.
Standard Analysis Package includes standard output of cellRanger software:
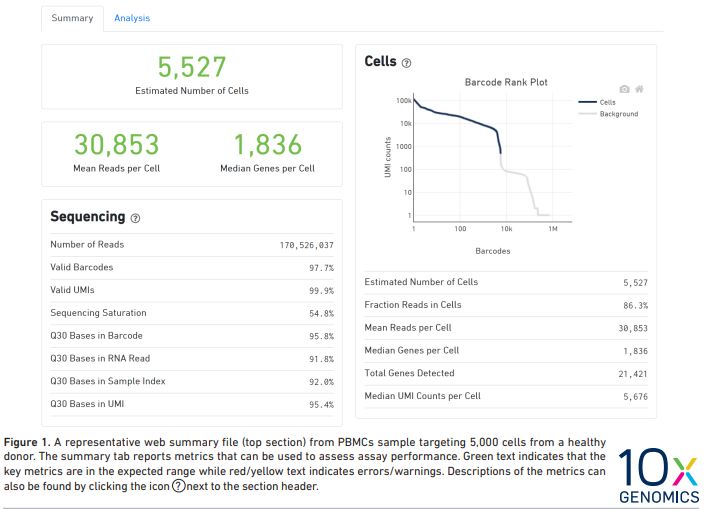
(1) Sample Web Summaries
(2) Counts data
(3) Raw data (fastq files)
The counts data can then be easily read into other downstream analytical tools like Loupe Browser and Seurat .
Example Web Summary Report:
10X Genomics Web summary Interpretation Document
ChIP/ATAC-Seq
Sequencing design= ChIP-Seq: 1x100; ATAC-Seq: 2x75
Sequencing platform = NextSeq2000/NovaSeq 6000/NovaSeq X
This pipeline is designed to work with data generated for ChIP-Seq and ATAC-Seq experiments.
Standard Analysis Package Includes:
(1) Continuous coverage data in the bigwig and bedgraph format
(2) Enrichments in the narrowPeak, broadPeak, or BED format
We can also discuss analysis plans for downstream motif enrichment, differential binding, nucelosomal positioning, and nearest gene annotation in a consulting meeting since these types of analyses tend to change based on the underlying hypothesis.
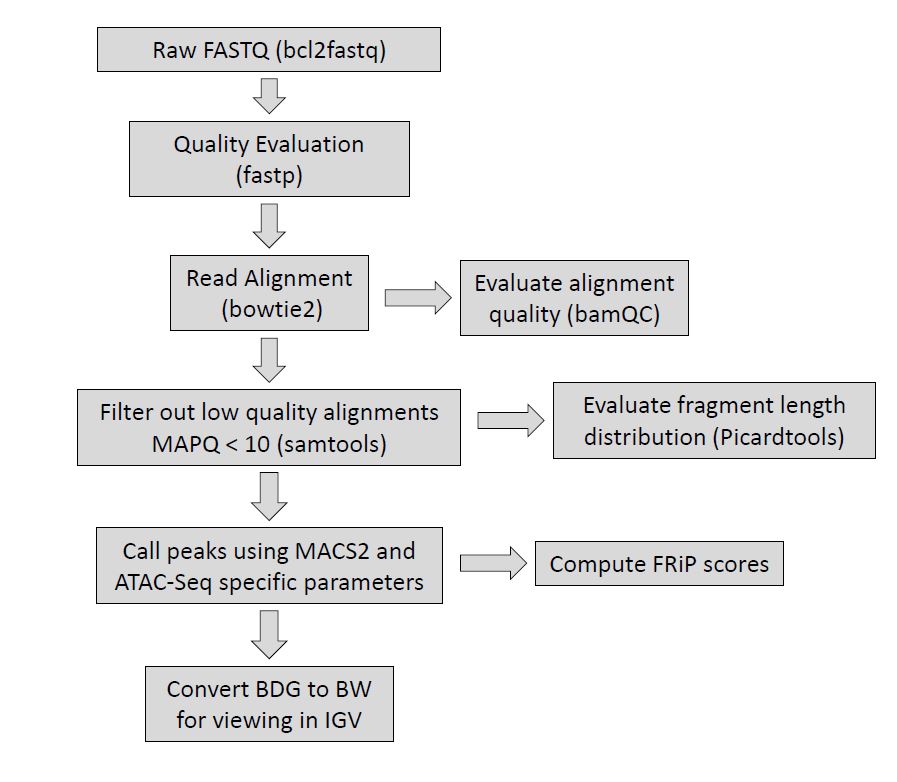
The current software that we use to generate our preliminary results: fastp, bowtie2, deeptools alignment sieve, samtools, bamqc, picardtools, macs2 (with project specific parameters).
Figure 2. Overview of the Epigenetics Pipeline specifically for ATAC-Seq, indicating what tools are used at each step. Please note, the fragment length distribution computed by picardtools will also only be computed for paired-end approaches.
Microbiome
16S rRNA
Sequencing design= 2x300 (V3-V4 & V1-V3); 2x150 (V4)
Sequencing platform = MiSeq/NextSeq 2000
16S rRNA hypervariable regions V1-V3 or V3-V4. Primary processing using QIIME 2 including primer removal and end trimming. Forward and reverse read merging, chimera removal, quality filtering, and denoising with DADA2. Taxonomic classification with target region-specific naive Bayesian classifier trained on the GreenGenes or SLIVA reference databases.
Standard Analysis Package Includes:
(1) Sequences of amplicon variants
(2) Taxonomic assignments to sequence variants
(3) Associated counts.
These can be generated at various taxonomic resolutions (e.g. species, genus, etc.)
Shotgun Metagenomics or Metatranscriptomics
Sequencing Platform = please inquire. Will vary based on sample number and type.
Reads are preprocessed to remove Illumina adapters, low quality bases, and host/rRNA contaminants. Taxonomic and function profiling can be performed using read-based and/or assembly-based methods. Our read-based workflow uses Metaphlan and Humman from the BioBakery suite of tools developed by the Huttenhower lab. This approach maps reads to a taxonomic and functional marker gene database and is a relatively fast way to profile communities with relevant reference genomes, primarily the human gut microbiome. Our assembly-based workflow performs de-novo assembly of reads into contigs and groups contigs into bins based on sequence similarity. These bins are then given a taxonomic assignment based on homology to the NCBI nt database. Genes are called within these bins and then assigned functions from multiple protein and metabolic databases like PFAM and KEGG. The assembly-based workflow into for is ideal for non-human samples or projects seeking strain level resolution for phylogenetics and comparative genomics.
Standard Analysis Package Includes:
(1) Tables of taxonomic, gene, and metabolic pathway abundance
(2) Sequences for all assembled contigs/bins
(3) Metrics from strain level genome comparisons (SNPs and SNP linkage, contig coverage, and contig homology)
Whole Genome & Whole Exome Sequencing (WGS/WES)
Sequencing design= 2x150
Sequencing platform = NovaSeq 6000/NovaSeq X
The GATK best practices pipeline is used to align WGS and WES data to the human reference genome (GRCh38/hg38), call SNPs and INDELs, filter variants to reduce false positives, and annotate with known information about each loci and potential functional consequences. Results deliverables include: all variant calls with annotations in VCF format.
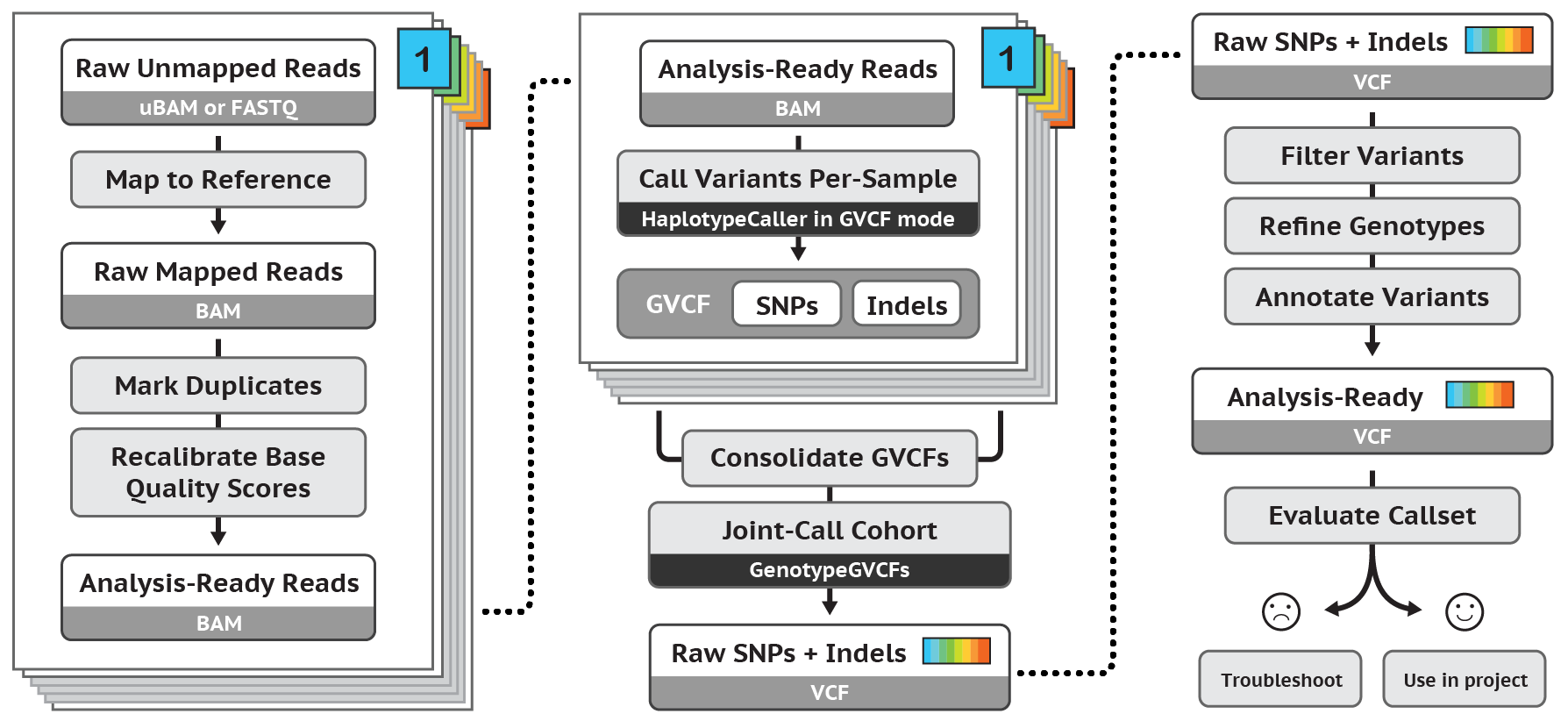
Figure 3. Overview of GATK pipeline reference: https://software.broadinstitute.org/gatk/