GRC Sequencing Instrumentation

New Addition: Illumina NovaSeq X Plus Sequencer
Learn more about the NovaSeq X system

The NovaSeq X has patterned flow cell technology and it is highly recommended that samples utilize the Unique Dual Indexing strategy to minimize the risk of index hopping.
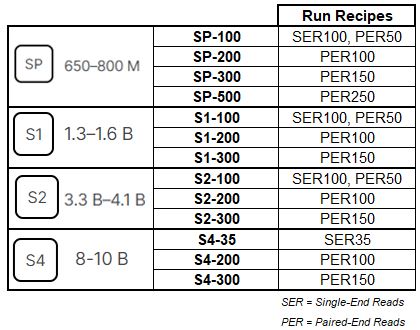
NovaSeq X Performance Specifications
Click here to see a technical note about RNAseq performance between the NovaSeq X and the NovaSeq 6000.
NovaSeq Applications:
- Whole Genome Sequencing
- Whole Exome Sequencing
- Whole Transcriptome Sequencing
*Note: The GRC reserves the right to determine the most appropriate sequencing instrument for your project and will proceed accordingly. If there is a specific sequencer you would like to utilize for your project, please indicate this on your submission sheet.
Illumina NovaSeq 6000 Sequencer
Learn more about the NovaSeq6000 system

The NovaSeq 6000 has patterned flow cell technology and it is highly recommended that samples utilize the Unique Dual Indexing strategy to minimize the risk of index hopping.
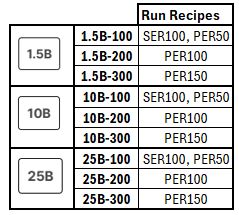
NovaSeq 6000 Performance Specifications
NovaSeq Applications:
- Whole Genome Sequencing
- Whole Exome Sequencing
- Whole Transcriptome Sequencing
New Addition: Illumina NextSeq 2000
Learn more about the NextSeq 2000 System
The NextSeq 2000 is the latest addition to sequencing instruments at the GRC. This instrument will replace the NextSeq 550 (Jan 1, 2024) and provide researchers with greater flexibility, faster run times, and lower sequencing costs.
The NextSeq 2000 has patterned flow cell technology and it is highly recommended that samples utilize the Unique Dual Indexing strategy to minimize the risk of index hopping.
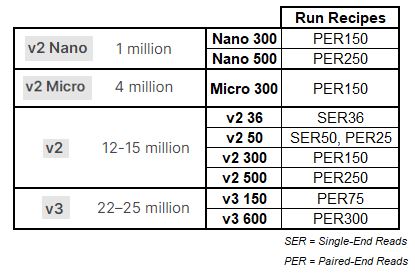
NextSeq 2000 Performance Specifications
NextSeq 2000 Applications:
- Exome Sequencing
- Whole Transcriptome Sequencing
- Small Whole Genome Sequencing
- Epigenetic Sequencing
Illumina NextSeq 550 (Sunset: January 1, 2024)
Learn more about the NextSeq 550 System 
NextSeq 550 Performance Specifications
NextSeq 550 Applications:
- Gene Expression Profiling
- Exome Sequencing
- Whole Genome Sequencing
Announcement regarding sunset of NextSeq 550 (January 1, 2024)
As of January 1, 2024, we will no longer be sequencing with the NextSeq 550. This platform has been used for projects that have not yet incorporated the Unique Dual Indexing (UDI) strategy. If your lab needs assistance with modification of lab protocols to incorporate UDIs, please contact us for assistance. Projects that utilize alternative indexing strategies (i.e. single index or combinatorial dual) can still be sequenced on all platforms with the understanding that index hopping may occur.
Illumina MiSeq
Learn more about the MiSeq System
MiSeq performance specifications
MiSeq Applications:
- Amplicon Sequencing
- 16s (microbiome)
- Targeted/Custom Panels
- Rep-seq
- Targeted Amplicon Panels
- TruSeq Cancer Amplicon (LINK)
- TruSight Myeloid Panel (LINK)
- Small Genome Sequencing
- Bacteria, Viruses, etc.
Recommended Reads Per Library
| Application | Reads per Library |
|---|---|
| mRNA-Seq | 20 - 25 Million |
| Total RNA-Seq | 40 - 50 Million |
| Small RNA-Seq | 2 Million |
| Low Input RNA-Seq | 20 - 25 Million |
| Chip-Seq | 15 Million |
| ATAC-Seq | 50 Million |
| Whole Genome (10X) | 125 Million Pairs |
| Whole Genome (4X) | 50 Million Pairs |
| Whole Genome Bisulfite-Seq (10X) | 125 Million Pairs |
| Whole Exome Sequencing (100X) | 50 Million Pairs |
| Single Cell (10X v3) | 100,000 per Cell |
**The above recommendations are based on Human/Mouse/Rat genomes for simplicity and empirical results from GRC experience. Other organisms may require different sequencing depths (more or less samples per lane). The amount of sequencing performed and the type (single vs pair) is organism and application specific. These recommendations are guidelines and by no means meant to be strict cutoffs. Internal GRC data and cumulative data analysis over the past formed the basis for these suggestions with the sole purpose to help investigators select the most appropriate experiment type for their research goals.
Contact us or use the Sequencing Platform Comparison Tool to find the right alternative for your research needs.
Data Analysis
The GRC has an experienced group of bioinformatics staff with experience in NGS data analysis methods and workflows. For more information please visit the GRC Bioinformatics Page or contact us.